
|
Central asymmetric unit spacefilled. |
|
Conformation May Be Altered by Crystal Contacts
Characteristics of Crystal Contacts vs. Oligomer Interfaces Visualizing Crystal Contacts in Protein Explorer References |
The term crystal contacts means interchain or intermolecular contacts that occur solely as the result of protein crystallization (2). These are distinguished from oligomer interfaces that occur in the uncrystallized protein. Both oligomer interfaces and crystal contacts typically occur in crystals, but only the latter are artifacts of crystallization. For more about oligomers, and their structures, see

|
Central asymmetric unit spacefilled. |

|
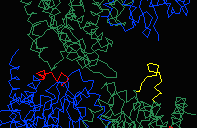 In 4MDH, each chain in the homodimer experiences different crystal contacts.
Chain A has crystal contacts near the catalytic site, absent for chain B.
These crystal contacts influence the conformation of residues 93-99.
To visualize this in Chime, see
Crystal Contacts in Malate Dehydrogenase. (Thanks to
Gale Rhodes for this example.)
In 4MDH, each chain in the homodimer experiences different crystal contacts.
Chain A has crystal contacts near the catalytic site, absent for chain B.
These crystal contacts influence the conformation of residues 93-99.
To visualize this in Chime, see
Crystal Contacts in Malate Dehydrogenase. (Thanks to
Gale Rhodes for this example.)

|
differing conformations of 93-99 (stereo). |
The asymmetric unit in 4MDH (contained in the published PDB file available from the Protein Data Bank) contains one homodimer. The crystallographic unit cell contains 4 homodimers. The above-linked Chime presentation on 4MDH displays three adjacent crystallographic unit cells generated with DeepView (figure at left). In all three figures above, residues 93-99 are colored red or yellow. The yellow loops are surrounded by solvent, while the red loops contact another chain in the crystal. This contact appears to be responsible for the different conformations of the loops.
It is also possible that a protein can have two different
conformations in two different crystals, resulting from
the occurrence of differing crystal contacts in each case.
(A good example of this would be welcome.)
Dasgupta et al. (2) studied 58 oligomers and 223 nonvirus crystals with good resolution and completeness. They found that "crystal contact patches are frequently smaller than patches involved in oligomer interfaces". Contact patches of 10 to 100 atoms were common at crystal contacts. Patches involving 100-1,000 atoms were common in oligomer interfaces but rarely seen in crystal contacts. Nevertheless, the total number of atoms involved in crystal contacts vs. oligomer interfaces were about the same; that is, the larger number of small crystal contact patches involved about the same number of atoms as the smaller number of large oligomer interfaces. They also observed that crystal contacts tend to involve more polar interactions, while nonpolar interactions tend to predominate in oligomer interfaces. "Hydrophobic interactions lead to disordered precipitates, and not to crystals."
Published PDB files never show all of the crystal contacts occurring on each chain. It is a good idea to look at all contacts for each chain to get an idea where they might be affecting the conformation. Unfortunately, there is presently no server that generates a PDB file ready for inspection of crystal contacts in Protein Explorer.
Here are some examples of PDB files showing crystal contacts. These files were constructed using the DeepView/RasMol method described below. Each is divided into two models delimited as "NMR" models. Model 1 is the original aysmmetric unit, and model 2 contains all crystal contact atoms within 5 Å.
162-molecule, 27-unit cell "crystal":
Gert Vriend of the Centre for Molecular and Biomolecular Informatics, Katholieke Universiteit Nijmegen, is working on a crystal contacts server, part of the WHAT IF WWW Interface. The present version can be found there under Crystal Symmetry, Add shell of symmetry related residues. However, it is not finished and the results are not guaranteed to be correct, nor is the resulting PDB file very compatible with Protein Explorer.
At present, the best way to view crystal contacts is to generate them with DeepView. This is a somewhat laborious process, despite the many timesaving features of this program. Here are instructions on how to generate crystal contacts with DeepView. In the resulting PDB file, it is difficult to visualize the contacts to the asymmetric unit because it is buried in the center of many chains. But from this file, a subset can be saved containing the asymmetric unit (or a single chain) plus only a shell of all contacting atoms within a specified distance, such as 5 Å (explained in the above instructions).
|
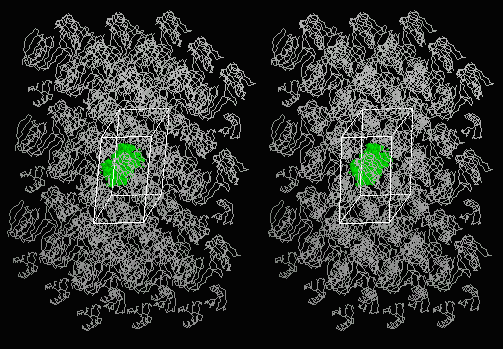
Below: Stereo view of "crystal" of 1PGB with central asymmetric unit in green. |
|